Bioinfo-Assignment-1
I. Assembly and de Bruijn graph. Given the following reads, draw the de Bruijn graph (3-mer as node), and write down what is the whole sequence.
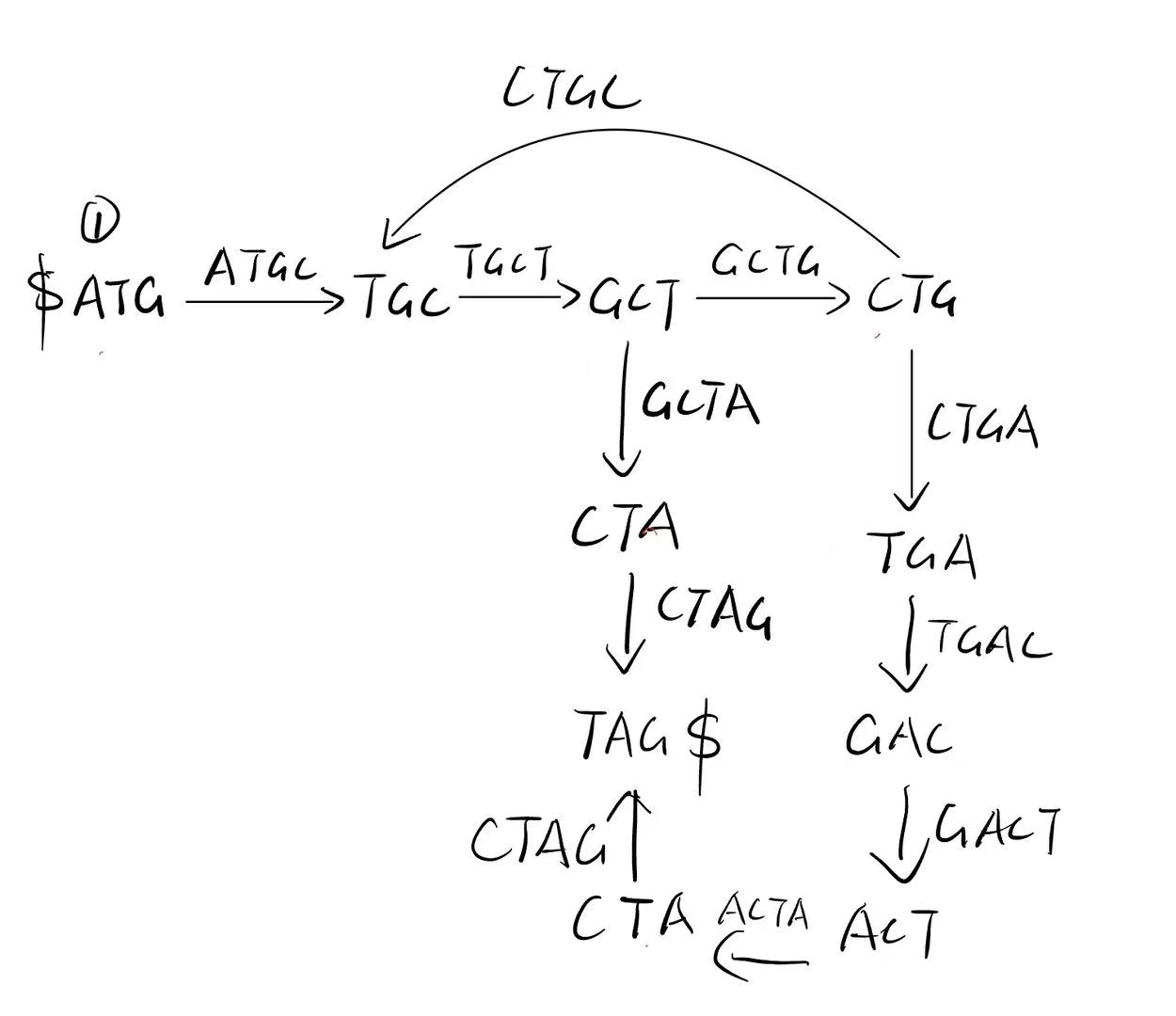
Path:
- ATGCTAG
- ATGCTGACTAG
2. Here is the position and weight of all exon segments in an abstract sequence of length 18. Please find a maximum set of non-overlapping putative exons based on brute force or dynamic programming.
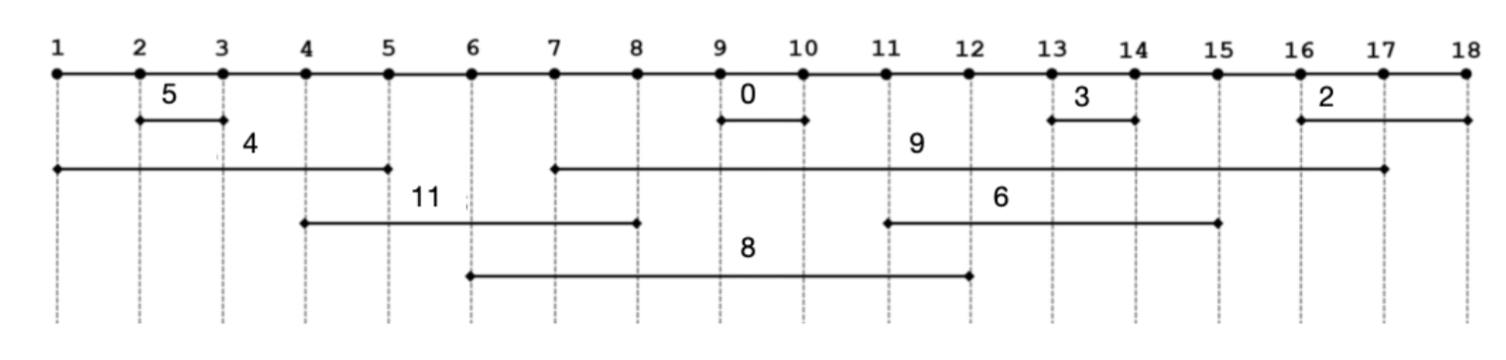
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 5 | 5 | 5 | 5 | 5 | 16 | 16 | 16 | 16 | 16 | 16 | 19 | 22 | 22 | 22 | 24 |
Selected exons:
| left | right | weight |
|---|---|---|
| 2 | 3 | 5 |
| 4 | 8 | 11 |
| 9 | 10 | 0 |
| 11 | 15 | 6 |
| 16 | 18 | 2 |
3. Please generate a dynamic programming matrix for sequence alignment and find its optimum alignment score.
| G | A | T | C | A | G | A | T | C | G | A | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -6 | -12 | -18 | -24 | -30 | -36 | -42 | -48 | -54 | -60 | -66 | ||
| G | -6 | -1 | -7 | -13 | -19 | -25 | -31 | -37 | -43 | -49 | -55 | |
| G | -12 | -1 | -3 | -9 | -15 | -14 | -20 | -26 | -32 | -38 | -44 | |
| T | -18 | -7 | -3 | 2 | -4 | -10 | -16 | -15 | -21 | -27 | -33 | |
| A | -24 | -13 | -2 | 2 | 1 | -5 | -11 | -17 | -23 | -22 | ||
| A | -30 | -19 | -8 | -4 | 0 | 0 | -6 | -12 | -18 | |||
| C | -36 | -25 | -14 | -10 | 1 | 5 | 9 | 3 | -1 | -7 | ||
| G | -42 | -31 | -20 | -16 | -5 | -1 | 10 | 7 | 1 | 2 | 4 | |
| T | -48 | -37 | -26 | -15 | -11 | -7 | 4 | 8 | 12 | 6 | 4 |
4. To apply database/resource and tools
Search genes associated with colon cancer in human genome.
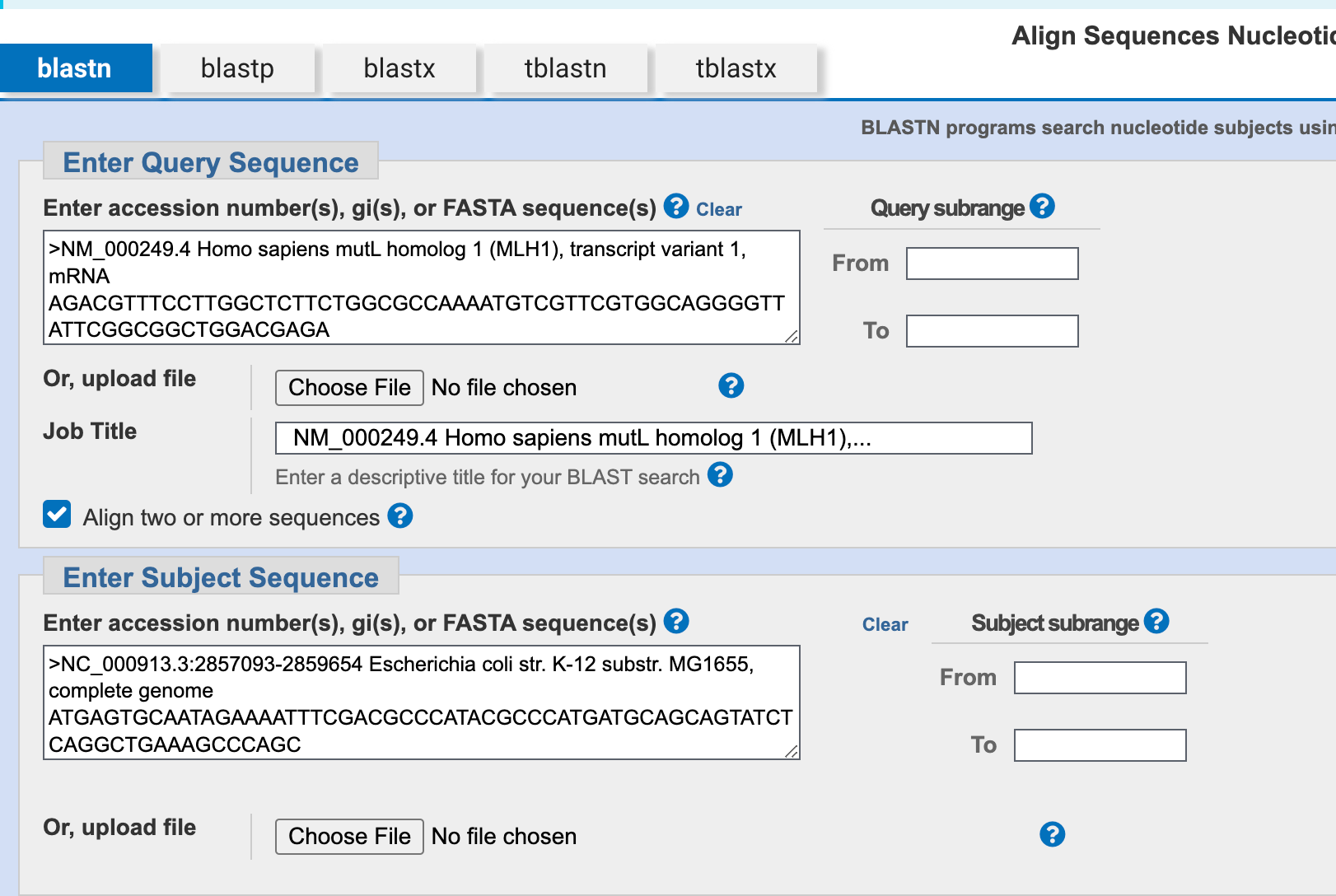
1. Give the DNA sequence of MLH1 (nucleotide NM_000249).
fasta
>NM_000249.4 Homo sapiens mutL homolog 1 (MLH1), transcript variant 1, mRNA
AGACGTTTCCTTGGCTCTTCTGGCGCCAAAATGTCGTTCGTGGCAGGGGTTATTCGGCGGCTGGACGAGA
CAGTGGTGAACCGCATCGCGGCGGGGGAAGTTATCCAGCGGCCAGCTAATGCTATCAAAGAGATGATTGA
GAACTGTTTAGATGCAAAATCCACAAGTATTCAAGTGATTGTTAAAGAGGGAGGCCTGAAGTTGATTCAG
ATCCAAGACAATGGCACCGGGATCAGGAAAGAAGATCTGGATATTGTATGTGAAAGGTTCACTACTAGTA
AACTGCAGTCCTTTGAGGATTTAGCCAGTATTTCTACCTATGGCTTTCGAGGTGAGGCTTTGGCCAGCAT
AAGCCATGTGGCTCATGTTACTATTACAACGAAAACAGCTGATGGAAAGTGTGCATACAGAGCAAGTTAC
TCAGATGGAAAACTGAAAGCCCCTCCTAAACCATGTGCTGGCAATCAAGGGACCCAGATCACGGTGGAGG
ACCTTTTTTACAACATAGCCACGAGGAGAAAAGCTTTAAAAAATCCAAGTGAAGAATATGGGAAAATTTT
GGAAGTTGTTGGCAGGTATTCAGTACACAATGCAGGCATTAGTTTCTCAGTTAAAAAACAAGGAGAGACA
GTAGCTGATGTTAGGACACTACCCAATGCCTCAACCGTGGACAATATTCGCTCCATCTTTGGAAATGCTG
TTAGTCGAGAACTGATAGAAATTGGATGTGAGGATAAAACCCTAGCCTTCAAAATGAATGGTTACATATC
CAATGCAAACTACTCAGTGAAGAAGTGCATCTTCTTACTCTTCATCAACCATCGTCTGGTAGAATCAACT
TCCTTGAGAAAAGCCATAGAAACAGTGTATGCAGCCTATTTGCCCAAAAACACACACCCATTCCTGTACC
TCAGTTTAGAAATCAGTCCCCAGAATGTGGATGTTAATGTGCACCCCACAAAGCATGAAGTTCACTTCCT
GCACGAGGAGAGCATCCTGGAGCGGGTGCAGCAGCACATCGAGAGCAAGCTCCTGGGCTCCAATTCCTCC
AGGATGTACTTCACCCAGACTTTGCTACCAGGACTTGCTGGCCCCTCTGGGGAGATGGTTAAATCCACAA
CAAGTCTGACCTCGTCTTCTACTTCTGGAAGTAGTGATAAGGTCTATGCCCACCAGATGGTTCGTACAGA
TTCCCGGGAACAGAAGCTTGATGCATTTCTGCAGCCTCTGAGCAAACCCCTGTCCAGTCAGCCCCAGGCC
ATTGTCACAGAGGATAAGACAGATATTTCTAGTGGCAGGGCTAGGCAGCAAGATGAGGAGATGCTTGAAC
TCCCAGCCCCTGCTGAAGTGGCTGCCAAAAATCAGAGCTTGGAGGGGGATACAACAAAGGGGACTTCAGA
AATGTCAGAGAAGAGAGGACCTACTTCCAGCAACCCCAGAAAGAGACATCGGGAAGATTCTGATGTGGAA
ATGGTGGAAGATGATTCCCGAAAGGAAATGACTGCAGCTTGTACCCCCCGGAGAAGGATCATTAACCTCA
CTAGTGTTTTGAGTCTCCAGGAAGAAATTAATGAGCAGGGACATGAGGTTCTCCGGGAGATGTTGCATAA
CCACTCCTTCGTGGGCTGTGTGAATCCTCAGTGGGCCTTGGCACAGCATCAAACCAAGTTATACCTTCTC
AACACCACCAAGCTTAGTGAAGAACTGTTCTACCAGATACTCATTTATGATTTTGCCAATTTTGGTGTTC
TCAGGTTATCGGAGCCAGCACCGCTCTTTGACCTTGCCATGCTTGCCTTAGATAGTCCAGAGAGTGGCTG
GACAGAGGAAGATGGTCCCAAAGAAGGACTTGCTGAATACATTGTTGAGTTTCTGAAGAAGAAGGCTGAG
ATGCTTGCAGACTATTTCTCTTTGGAAATTGATGAGGAAGGGAACCTGATTGGATTACCCCTTCTGATTG
ACAACTATGTGCCCCCTTTGGAGGGACTGCCTATCTTCATTCTTCGACTAGCCACTGAGGTGAATTGGGA
CGAAGAAAAGGAATGTTTTGAAAGCCTCAGTAAAGAATGCGCTATGTTCTATTCCATCCGGAAGCAGTAC
ATATCTGAGGAGTCGACCCTCTCAGGCCAGCAGAGTGAAGTGCCTGGCTCCATTCCAAACTCCTGGAAGT
GGACTGTGGAACACATTGTCTATAAAGCCTTGCGCTCACACATTCTGCCTCCTAAACATTTCACAGAAGA
TGGAAATATCCTGCAGCTTGCTAACCTGCCTGATCTATACAAAGTCTTTGAGAGGTGTTAAATATGGTTA
TTTATGCACTGTGGGATGTGTTCTTCTTTCTCTGTATTCCGATACAAAGTGTTGTATCAAAGTGTGATAT
ACAAAGTGTACCAACATAAGTGTTGGTAGCACTTAAGACTTATACTTGCCTTCTGATAGTATTCCTTTAT
ACACAGTGGATTGATTATAAATAAATAGATGTGTCTTAACATAA2. Give the DNA sequence of E. coli mismatch repair gene mutS.
DNA Sequence of E. coli mismatch repair gene mutS
fasta
>NC_000913.3:2857093-2859654 Escherichia coli str. K-12 substr. MG1655, complete genome
ATGAGTGCAATAGAAAATTTCGACGCCCATACGCCCATGATGCAGCAGTATCTCAGGCTGAAAGCCCAGC
ATCCCGAGATCCTGCTGTTTTACCGGATGGGTGATTTTTATGAACTGTTTTATGACGACGCAAAACGCGC
GTCGCAACTGCTGGATATTTCACTGACCAAACGCGGTGCTTCGGCGGGAGAGCCGATCCCGATGGCGGGG
ATTCCCTACCATGCGGTGGAAAACTATCTCGCCAAACTGGTGAATCAGGGAGAGTCCGTTGCCATCTGCG
AACAAATTGGCGATCCGGCGACCAGCAAAGGTCCGGTTGAGCGCAAAGTTGTGCGTATCGTTACGCCAGG
CACCATCAGCGATGAAGCCCTGTTGCAGGAGCGTCAGGACAACCTGCTGGCGGCTATCTGGCAGGACAGC
AAAGGTTTCGGCTACGCGACGCTGGATATCAGTTCCGGGCGTTTTCGCCTGAGCGAACCGGCTGACCGCG
AAACGATGGCGGCAGAACTGCAACGCACTAATCCTGCGGAACTGCTGTATGCAGAAGATTTTGCTGAAAT
GTCGTTAATTGAAGGCCGTCGCGGCCTGCGCCGTCGCCCGCTGTGGGAGTTTGAAATCGACACCGCGCGC
CAGCAGTTGAATCTGCAATTTGGGACCCGCGATCTGGTCGGTTTTGGCGTCGAGAACGCGCCGCGCGGAC
TTTGTGCTGCCGGTTGTCTGTTGCAGTATGCGAAAGATACCCAACGTACGACTCTGCCGCATATTCGTTC
CATCACCATGGAACGTGAGCAGGACAGCATCATTATGGATGCCGCGACGCGTCGTAATCTGGAAATCACC
CAGAACCTGGCGGGTGGTGCGGAAAATACGCTGGCTTCTGTGCTCGACTGCACCGTCACGCCGATGGGCA
GCCGTATGCTGAAACGCTGGCTGCATATGCCAGTGCGCGATACCCGCGTGTTGCTTGAGCGCCAGCAAAC
TATTGGCGCATTGCAGGATTTCACCGCCGGGCTACAGCCGGTACTGCGTCAGGTCGGCGACCTGGAACGT
ATTCTGGCACGTCTGGCTTTACGAACTGCTCGCCCACGCGATCTGGCCCGTATGCGCCACGCTTTCCAGC
AACTGCCGGAGCTGCGTGCGCAGTTAGAAACTGTCGATAGTGCACCGGTACAGGCGCTACGTGAGAAGAT
GGGCGAGTTTGCCGAGCTGCGCGATCTGCTGGAGCGAGCAATCATCGACACACCGCCGGTGCTGGTACGC
GACGGTGGTGTTATCGCATCGGGCTATAACGAAGAGCTGGATGAGTGGCGCGCGCTGGCTGACGGCGCGA
CCGATTATCTGGAGCGTCTGGAAGTCCGCGAGCGTGAACGTACCGGCCTGGACACGCTGAAAGTTGGCTT
TAATGCGGTGCACGGCTACTACATTCAAATCAGCCGTGGGCAAAGCCATCTGGCACCCATCAACTACATG
CGTCGCCAGACGCTGAAAAACGCCGAGCGCTACATCATTCCAGAGCTAAAAGAGTACGAAGATAAAGTTC
TCACCTCAAAAGGCAAAGCACTGGCACTGGAAAAACAGCTTTATGAAGAGCTGTTCGACCTGCTGTTGCC
GCATCTGGAAGCGTTGCAACAGAGCGCGAGCGCGCTGGCGGAACTCGACGTGCTGGTTAACCTGGCGGAA
CGGGCCTATACCCTGAACTACACCTGCCCGACCTTCATTGATAAACCGGGCATTCGCATTACCGAAGGTC
GCCATCCGGTAGTTGAACAAGTACTGAATGAGCCATTTATCGCCAACCCGCTGAATCTGTCGCCGCAGCG
CCGCATGTTGATCATCACCGGTCCGAACATGGGCGGTAAAAGTACCTATATGCGCCAGACCGCACTGATT
GCGCTGATGGCCTACATCGGCAGCTATGTACCGGCACAAAAAGTCGAGATTGGACCTATCGATCGCATCT
TTACCCGCGTAGGCGCGGCAGATGACCTGGCGTCCGGGCGCTCAACCTTTATGGTGGAGATGACTGAAAC
CGCCAATATTTTACATAACGCCACCGAATACAGTCTGGTGTTAATGGATGAGATCGGGCGTGGAACGTCC
ACCTACGATGGTCTGTCGCTGGCGTGGGCGTGCGCGGAAAATCTGGCGAATAAGATTAAGGCATTGACGT
TATTTGCTACCCACTATTTCGAGCTGACCCAGTTACCGGAGAAAATGGAAGGCGTCGCTAACGTGCATCT
CGATGCACTGGAGCACGGCGACACCATTGCCTTTATGCACAGCGTGCAGGATGGCGCGGCGAGCAAAAGC
TACGGCCTGGCGGTTGCAGCTCTGGCAGGCGTGCCAAAAGAGGTTATTAAGCGCGCACGGCAAAAGCTGC
GTGAGCTGGAAAGCATTTCGCCGAACGCCGCCGCTACGCAAGTGGATGGTACGCAAATGTCTTTGCTGTC
AGTACCAGAAGAAACTTCGCCTGCGGTCGAAGCTCTGGAAAATCTTGATCCGGATTCACTCACCCCGCGT
CAGGCGCTGGAGTGGATTTATCGCTTGAAGAGCCTGGTGTAA3. Compare MLH1 and mutS (answer of previous two questions) sequence.

4. Translate the above two gene sequences to protein sequences.
Human MLH1 Protein Sequence
txt
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERCL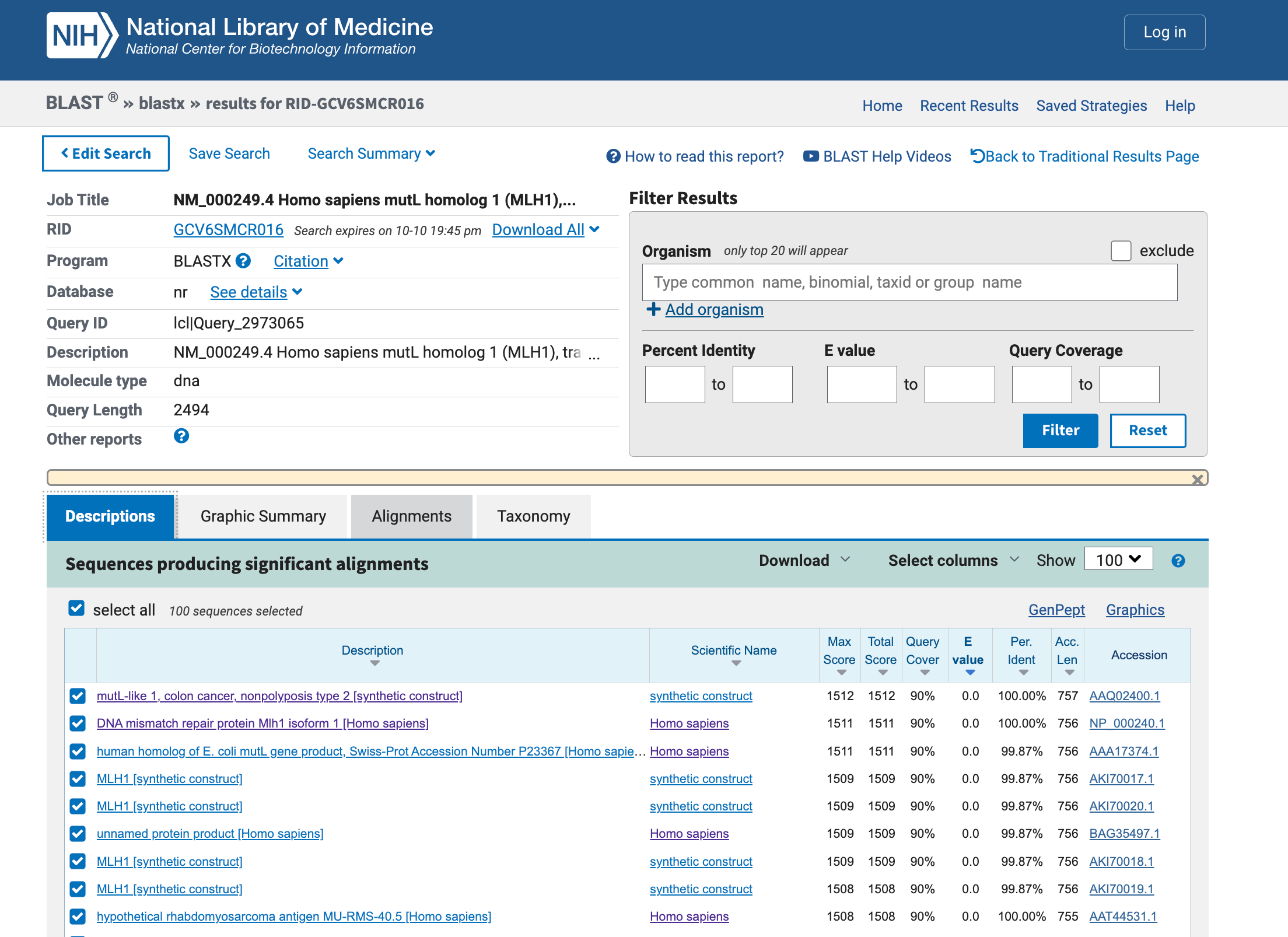
E. coli mismatch repair gene mutS Protein Sequence
txt
MSAIENFDAHTPMMQQYLRLKAQHPEILLFYRMGDFYELFYDDAKRASQLLDISLTKRGASAGEPIPMAGIPYHAVENYL
AKLVNQGESVAICEQIGDPATSKGPVERKVVRIVTPGTISDEALLQERQDNLLAAIWQDSKGFGYATLDISSGRFRLSEP
ADRETMAAELQRTNPAELLYAEDFAEMSLIEGRRGLRRHPLWEFEIDTARQQLNLQFGTRDLVGFGVENAPRGLCAAGCL
LQYAKDTQRTTLPHIRSITMEREQDSIIMDAATRRNLEITQNLAGGAENTLASVLDCTVTPMGSRMLKRWLHMPVRDTRV
LLERQQTIGALQDFTAGLQPVLRQVGDLERILARLALRTARPRDLARMRHAFQQLPELRAQLETVDSAPVQALREKMGEF
AELRDLLERAIIDTPPVLVRDGGVIASGYNEELDEWRALADGATDYLERLEVRERERTGLDTLKVGFNAVHGYYIQISRG
QSHLAPINYMRRQTLKNAERYIIPELKEYEDKVLTSKGKALALEKQLYEELFDLLLPHLEALQQSASALAELDVLVNLAE
RAYTLNYTCPTFIDKPGIRITEGRHPVVEQVLNEPFIANPLNLSPQRRMLIITGPNMGGKSTYMRQTALIALMAYIGSYV
PAQKVEIGPIDRIFTRVGAADDLASGRSTFMVEMTETANILHNATEYSLVLMDEIGRGTSTYDGLSLAWACAENLANKIK
ALTLFATHYFELTQLPEKMEGVANVHLDALEHGDTIAFMHSVQDGAASKSYGLAVAALAGVPKEVIKRARQKLRELESIS
PNAAATQVDGTQMSLLSVPEETSPAVEALENLDPDSLTPRQALEWIYRLKSLV
5. Perform protein sequence homology searching for MLH1 in GenBank. Give the 10 highest hits.
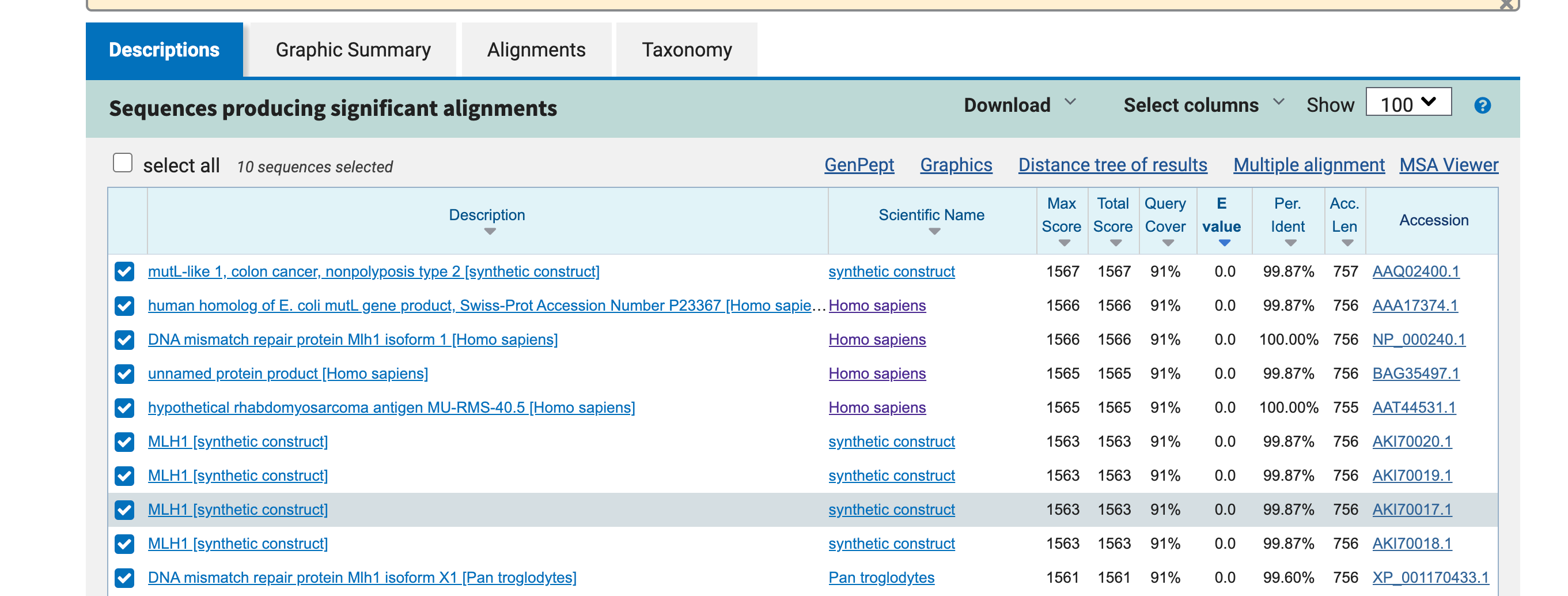
txt
>AAA17374.1 human homolog of E. coli mutL gene product, Swiss-Prot Accession Number P23367 [Homo sapiens]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSVFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>NP_000240.1 DNA mismatch repair protein Mlh1 isoform 1 [Homo sapiens]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>AAQ02400.1 mutL-like 1, colon cancer, nonpolyposis type 2, partial [synthetic construct]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERCL
>AKI70020.1 MLH1, partial [synthetic construct]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLDMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>BAG35497.1 unnamed protein product [Homo sapiens]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEVVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>AKI70018.1 MLH1, partial [synthetic construct]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNDYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>AAT44531.1 hypothetical rhabdomyosarcoma antigen MU-RMS-40.5, partial [Homo sapiens]
SFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERFT
TSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIAT
RRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAFK
MNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEES
ILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPLS
KPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVEM
VEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELFY
QILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLPL
LIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIVY
KALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>AKI70017.1 MLH1, partial [synthetic construct]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKFTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>AKI70019.1 MLH1, partial [synthetic construct]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKIHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
>XP_001170433.1 DNA mismatch repair protein Mlh1 isoform X1 [Pan troglodytes]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSIHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVTAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINERGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC5. Please finish one of the following problems.
Write a program to calculate similarity between any pairs of protein sequences in the given input file “APOBEC3A.fasta”. (global alignment, gap penalty and gap extend penalty=-5). You can download scoring matrix from here According to your program, which two sequences are most similar?
python
from Bio import SeqIO
def read_fasta(file):
sequences = []
for record in SeqIO.parse(file, "fasta"):
sequences.append(str(record.seq))
return sequences
sequences = read_fasta("APOBEC3A.fasta")
def load_blosum62(file):
blosum62 = {}
with open(file) as f:
lines = f.readlines()
amino_acids = lines[0].split()
for line in lines[1:]:
parts = line.split()
row_acid = parts[0]
scores = list(map(int, parts[1:]))
blosum62[row_acid] = dict(zip(amino_acids, scores))
return blosum62
def needleman_wunsch(seq1, seq2, gap_penalty, blosum62):
n = len(seq1)
m = len(seq2)
score_matrix = [[0] * (m + 1) for _ in range(n + 1)]
for i in range(1, n + 1):
score_matrix[i][0] = gap_penalty * i
for j in range(1, m + 1):
score_matrix[0][j] = gap_penalty * j
for i in range(1, n + 1):
for j in range(1, m + 1):
match = score_matrix[i-1][j-1] + blosum62[seq1[i-1]][seq2[j-1]]
delete = score_matrix[i-1][j] + gap_penalty
insert = score_matrix[i][j-1] + gap_penalty
score_matrix[i][j] = max(match, delete, insert)
return score_matrix[n][m]Get the highest similarity
python
blosum62 = load_blosum62("BLOSUM62.txt")
gap_penalty = -5
sequences = read_fasta("APOBEC3A.fasta")
ans=(0,1)
res=-1
for i in range(len(sequences)):
for j in range(i + 1, len(sequences)):
score = needleman_wunsch(sequences[i], sequences[j], gap_penalty, blosum62)
if score>res:
res=score
ans=(i,j)
# print(f"Similarity between sequence {i+1} and {j+1}: {score}")
print(f"The most similar sequences are {ans[0]+1} and {ans[1]+1} with a similarity score of {res}")Result
The most similar sequences are sequence APOBEC3A_Macaca_fascicularis and sequence APOBEC3A_Macaca_leonina with a similarity score of 1103

