Network Deconvolution|Network deconvolution as a general method to distinguish direct dependencies in networks
Assumptions
定义一个观测到的相似度矩阵,矩阵里的值可以是变量
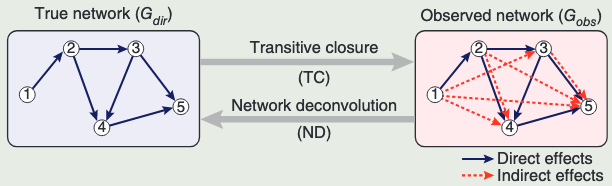
We assume that the observed dependency matrix,
观测矩阵包含直接和间接依赖效应
Indirect contributions
- Can be length 2 or higher
- Can be multiple effects along varying paths
- Not included Self-loops
间接影响可以是长度为2或更高,可以是沿着不同路径的多个效应,不包括自环
The power associated with each term in
Derivation
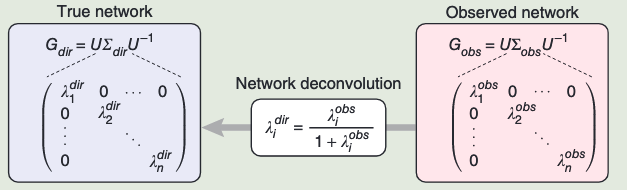
By using the eigen decomposition principle, we have
By using the eigen decomposition of the observed network
Network deconvolution Methods
Linear Scaling Step
The observed dependency matrix is scaled linearly so that all eigenvalues of the direct dependency matrix are between −1 and 1.
线性缩放使所有特征值在 -1~1 之间
Decomposition Step
The observed dependency matrix Gobs is decomposed to its eigenvalues and eigenvectors such that
Where:
is the matrix of eigenvectors, is a diagonal matrix containing the eigenvalues of , is the inverse of the eigenvector matrix .
将观察到的依赖矩阵分解为特征值和特征向量
Deconvolution Step
A diagonal eigenvalue matrix
Then, the output direct dependency matrix
根据公式计算
Experiments
Several network applications
- Distinguishing direct targets in gene expression regulatory networks
- 区分基因表达调控网络中的直接目标
- Recognizing directly interacting amino-acid residues for protein structure prediction from sequence alignments
- 通过序列比对识别直接相互作用的氨基酸残基以预测蛋白质结构
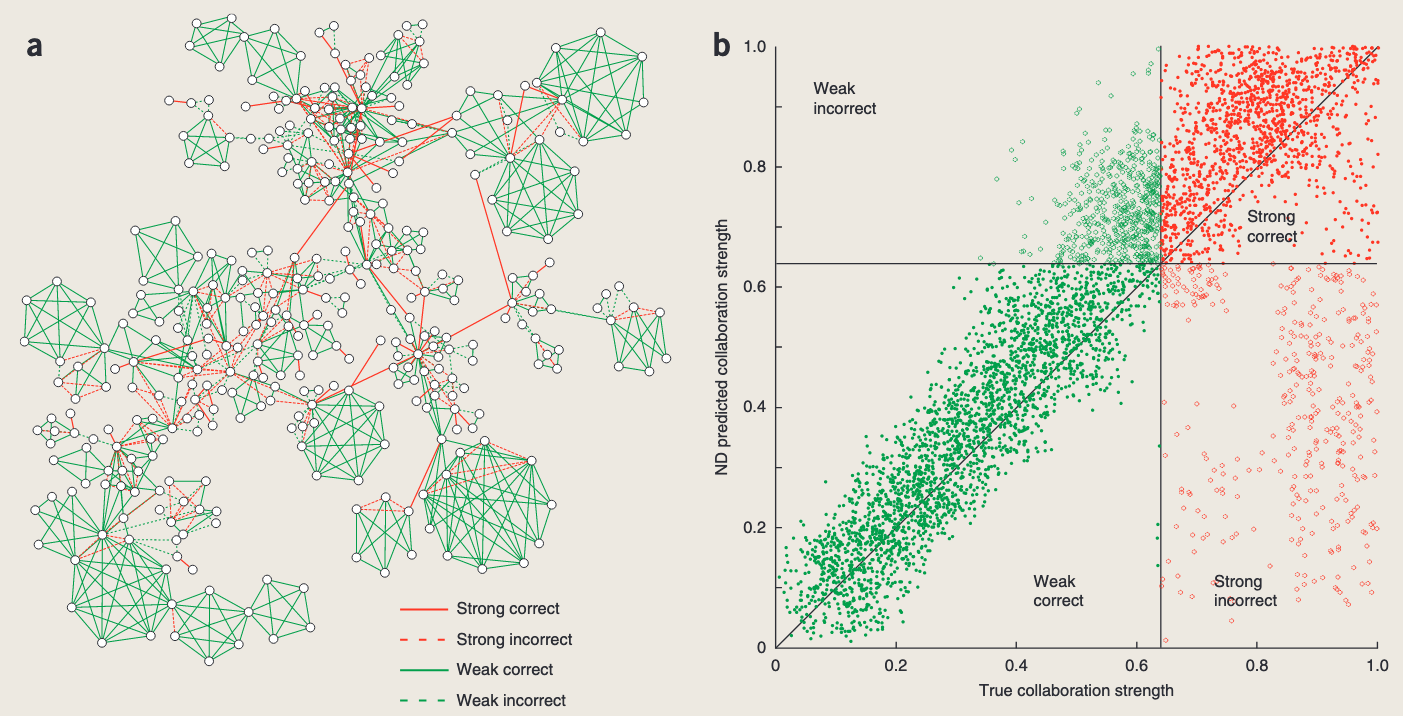
- Distinguishing strong collaborations in co-authorship social networks using connectivity information alone.
- 仅使用连接信息来区分共同创作社交网络中的强协作。
Gene expression regulatory networks
Distinguishing direct targets in gene expression regulatory networks
Social Network
Distinguishing strong collaborations in co-authorship social networks using connectivity information alone.